Analyzing Simulation Results
| Free Energy Fundamentals |
|---|
 |
|
Methods of Free Energy Simulations
|
| Free Energy How-to's |
|---|
 |
Once you have setup your transformation pathway, defined necessary intermediate states, run your equilibrium simulation, and acquired the correct data from the runs, you may now analyze the data to get free energies. Each of the free energy techniques discussed in the theory section of these fundamentals talks about how you get the free energies, but will also be recapped here. This page also talks about how you can get uncertainty estimates with the bootstrap method.
Calculating Free Energies
Shown below is a brief summary of the information needed and the method to calculating free energy with the various techniques. If you have read the pages for the free energy techniques, this will be a recap.
- EXP is a very straightforward calculation. You will need the [math]\displaystyle{ \Delta U }[/math] from a given pair of states. From there, you can calculate the free energy from the effective sample size found by correcting for correlation/subsampling. Variances will be additive.
- TI needs [math]\displaystyle{ \frac{dU}{d\lambda} }[/math] and the average at each of [math]\displaystyle{ K }[/math] states needs calculated. Since its more common to have discrete [math]\displaystyle{ \lambda }[/math] states, you will need to choose an appropriate weighting method to correctly calculate the free energy from the uncorrelated/subsampled data. By-state Variance will not be additive, but this is simple to account for.
- WHAM requires binning all your results then calculating [math]\displaystyle{ \Delta U }[/math] from all states. It is highly recommended that you take advantage of the tools already out there for calculating WHAM instead of writing your own. There is no direct way to calculate variance, and so methods such as bootstrap sampling are needed.
- BAR requires the [math]\displaystyle{ \Delta U }[/math] from two states. An iterative, often numeric, solution is needed to find the free energy, but this is relatively straightforward and can be done for each pair of states. The variance does not have as simple a relationship as with TI, and methods such as bootstrap sampling are recommended. There is a Python implementation of BAR available with PyMBAR.
- MBAR also requires [math]\displaystyle{ \Delta U }[/math], but it needs it for all combination of states. Again, an iterative set of solutions is needed and it is not recommended that users attempt to code MBAR themselves. Instead, please take advantage of a Python implementation available called PyMBAR. Error estimates are directly available with MBAR, however, it can still benefit from methods such as bootstrap sampling.
Bootstrap Sampling
Bootstrap sampling is a statistical tool in when we can get decent estimates for the uncertainty with very limited data;[1] this method works with all techniques presented in the fundamentals section. We carry out bootstrap sampling as the time taken to do bootstrap is often substantially less than the time it would take to generate new samples.
If we assume we have some function, [math]\displaystyle{ F(x) }[/math] that is computed from [math]\displaystyle{ N }[/math] data samples, [math]\displaystyle{ x_1,x_2\ldots,x_N }[/math], and we have that [math]\displaystyle{ \lim_{N\to\infty} F(x) = \overline{F} }[/math] where [math]\displaystyle{ \overline{F} }[/math] is a constant. As an example, say that [math]\displaystyle{ F(x) }[/math] is the simple average of all [math]\displaystyle{ x }[/math]
- [math]\displaystyle{ \displaystyle F(x)=\frac{1}{N}\sum\limits_{i=1}^N x_i }[/math],
or the average of some function [math]\displaystyle{ X(x) }[/math] such that
- [math]\displaystyle{ \displaystyle F(x)=\frac{1}{N}\sum\limits_{i=1}^N X(x_i) }[/math];
we are not limited to such simple choices as the functions could be as complicated the MBAR or WHAM free energy estimators. To calculate the bootstrap variance, carry out the following procedure.
- Pick with replacement [math]\displaystyle{ n }[/math] samples from the complete list of samples [math]\displaystyle{ \vec{x}\{x_1,\ldots,x_N\} }[/math] to create a new set of samples [math]\displaystyle{ \vec{x}_i }[/math]. Since you are picking randomly with replacement, there are bound to be repeated samples. This method is called sampling from the empirical distribution, that is, sampling from the distribution we measured, rather than true distribution. For example, if [math]\displaystyle{ \vec{x} = \{1,2,6,4,3\} }[/math], a possible set of [math]\displaystyle{ \vec{x}_i }[/math] might be [math]\displaystyle{ \vec{x}_i = \{6,1,1,4,4\} }[/math]; note that [math]\displaystyle{ \vec{x}_i }[/math] is the same size as the original [math]\displaystyle{ \vec{x} }[/math] set.
- Compute [math]\displaystyle{ \hat{F}_i = F(x_i) }[/math]. That is, find the estimate for your function based on the bootstrap sample taken from the empirical distribution. If we were finding simple averages, we would calculate the average of the bootstrap sample. If we were calculating free energies with MBAR, we would generate a bootstrap sample at each [math]\displaystyle{ K }[/math] state and estimate the full free energy at each state based on the bootstrap samples.
- Repeat steps 1 and 2 for [math]\displaystyle{ M }[/math] number of times. You will need at least 50-200 times to obtain accurate variances [1]. If the calculation in step 2 is cheap, then [math]\displaystyle{ M }[/math] can be very large to get even better estimates of the variance; relative uncertainty scales as [math]\displaystyle{ M^{-1/2} }[/math] and can take more than 1000 steps to get variance within 1% convergence.
- Compute the variance [math]\displaystyle{ \mathrm{var}\left(\hat{F}\right) }[/math], which is the variance over the [math]\displaystyle{ M }[/math] bootstrap values. The standard deviation is then just the square root of the bootstrap variance.
The values of [math]\displaystyle{ \hat{F_i} }[/math] are the bootstrap distribution of the function [math]\displaystyle{ F(x) }[/math]. In the large sample limit, the bootstrap distribution will converge to the true distribution of [math]\displaystyle{ \overline{F} }[/math] under most conditions, but will still be a good estimate for moderate values of [math]\displaystyle{ N }[/math]. If N is too low though, the bootstrap distribution will not have good agreement with the true distribution.
Bootstrap sampling can be used with any statistical estimator, regardless of how complicated. There is the extra overhead of calculating [math]\displaystyle{ F(x) }[/math] [math]\displaystyle{ M }[/math] times, but this is often negligible compared to how long it would take to collect [math]\displaystyle{ M }[/math] times the number of data. For MBAR, the time required for bootstrap is only 5-10min, where as TI is only seconds. There is also a variant of bootstrap called "moving block bootstrapping" which accounts for Simulation Information Gathering#Correlation time correlations without subsampling by taking random block of length [math]\displaystyle{ \tau }[/math].[1]
Example of Bootstrap Method
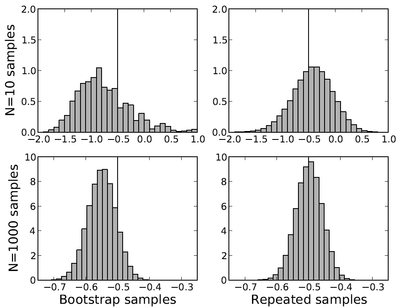
Lets take a simple example where we will sample values of [math]\displaystyle{ x }[/math] from a 1D Gaussian with mean 0 and and variance 1 ([math]\displaystyle{ x }[/math]~[math]\displaystyle{ N(0,1) }[/math]). We will now compute EXP with [math]\displaystyle{ k_BT=1 }[/math] for this sampling by
- [math]\displaystyle{ F(x_i) = -\ln \left[N^{-1}\sum_{i=1}^N \exp(-x_i)\right] }[/math].
We then compute the distribution of free energies obtained by [math]\displaystyle{ N }[/math] points for 100,000 bootstrap samples, and 100,000 independent samples (see figure on right). For small [math]\displaystyle{ N }[/math], the distributions are not close, but nearly converge for larger [math]\displaystyle{ N }[/math]. Even at N=200, the variances are near identical. To emphasize the point, the "Repeated Samples" distribution uses data from [math]\displaystyle{ 1000\times 100,000 }[/math] points collected, where the "Bootstrap Samples" needs only one set of 1000 samples run through the bootstrap process 100,000 times. The same amount of analysis is carried out in both cases, but the bootstrap samples only require 0.001% of the data collection. One catch to the bootstrap method that should be noted is that rare events requiring more than 1000 samples would not have been observed, so it still takes careful analysis to properly use bootstrap sampling.
References
- ↑ 1.0 1.1 1.2 Efron, B., and Tibshirani, R. J. An Introduction to the Bootstrap; Chapman and Hall/CRC:Boca Raton, FL, 1993. - Find at Cite-U-Like